Research
Our overarching goal is to explore computational modelling of quantitative MRI and validation techniques to establish new non-invasive biomarkers for cancer. An example is the VERDICT model, a biophysical model derived from a unique DWI acquisition protocol that aims to describe diffusion in tumours developed by Dr Eleftheria (Laura) Panagiotaki. It assumes that the diffusion contributions can be categorised into three primary components: vascular (isotropic restricted pseudo-diffusion), extracellular–extravascular space (hindered diffusion) and intracellular water (restricted diffusion).
VERDICT-MRI in the prostate
VERDICT-MRI has been shown to provide more microstructural feature information than images derived from similar models [1,2]. It was specifically designed to characterise prostate tissue, providing an accurate diagnosis from MRI and preventing unnecessary biopsies [3]. The model parameters have direct relevance to the tissue microstructure; these include intracellular volume faction (fIC), vascular volume fraction (fVASC), extracellular-extravascular volume fraction (fEES) and cell radius (R). The fIC and R maps can therefore be used to estimate cell density in the prostate and, consequently, detect PCa.
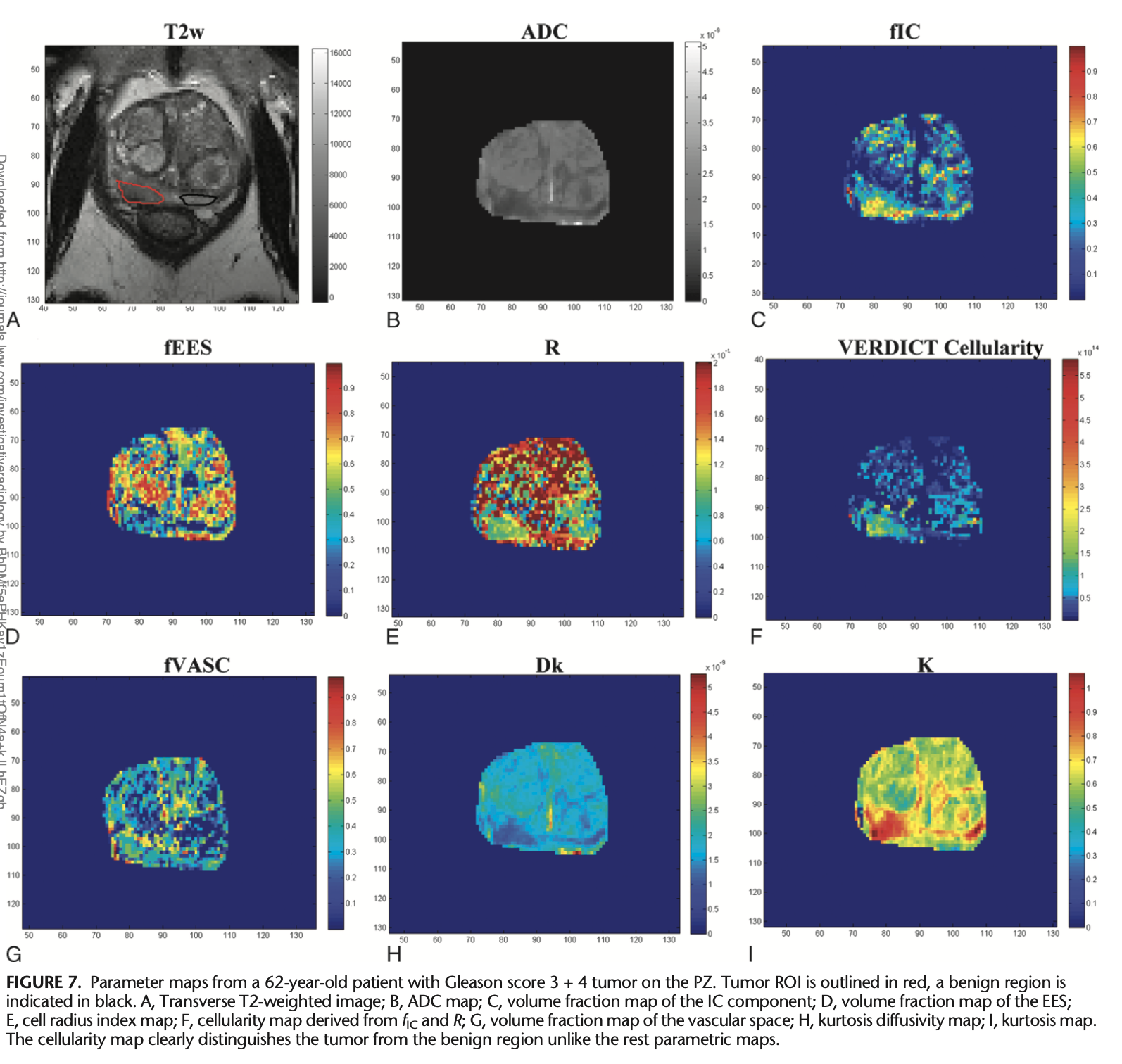
This work has been expanded to two main clinical trials that focus on validating VERDICT-MRI for PCa discrimination:
• INNOVATE [3] (April 2016 to December 2019): men suspected of having PCa were prospectively recruited from two centres and underwent VERDICT-MRI and mpMRI before undergoing targeted biopsy. The ADC, fIC, and prostate-specific antigen (PSA) density of the biopsied lesions were compared between men with csPCa and those without csPCa to test the diagnostic performance of each metric.
• Histo-MRI [4] (October 2020 to October 2023): consists of a prospectively recruited cohort of men suspected of having PCa. Histological analysis of men undergoing biopsy or prostatectomy is used for biological validation of biomarkers from VERDICT-MRI and Luminal Water Imaging that could help avoid unnecessary biopsies in men suspected of PCa.
This data has led to a series of studies that validate the VERDICT-MRI derived maps. An in-vivo validation of fIC, fEES and fVASC was presented in an abstract in 2017 [5]. Another abstract described the increased performance of fIC maps for discrimination between Gleason grades 3+3 and ≥ 3+4 when compared to ADC maps [6]. Johnston et al. also showed that fIC values outperformed ADC in differentiating Gleason 4 lesions from lower grade or healthy tissue [2], and they demonstrated high repeatability and image quality of the fIC maps. Chiou et al. showed the promise of using fully connected neural networks to classify VERDICT-MRI images into csPCa or non-csPCa [7]. Bailey et al. later investigated the effects of fixation on VERDICT parameters [8] using ex-vivo MRI, aligning the images through rigid registrations.
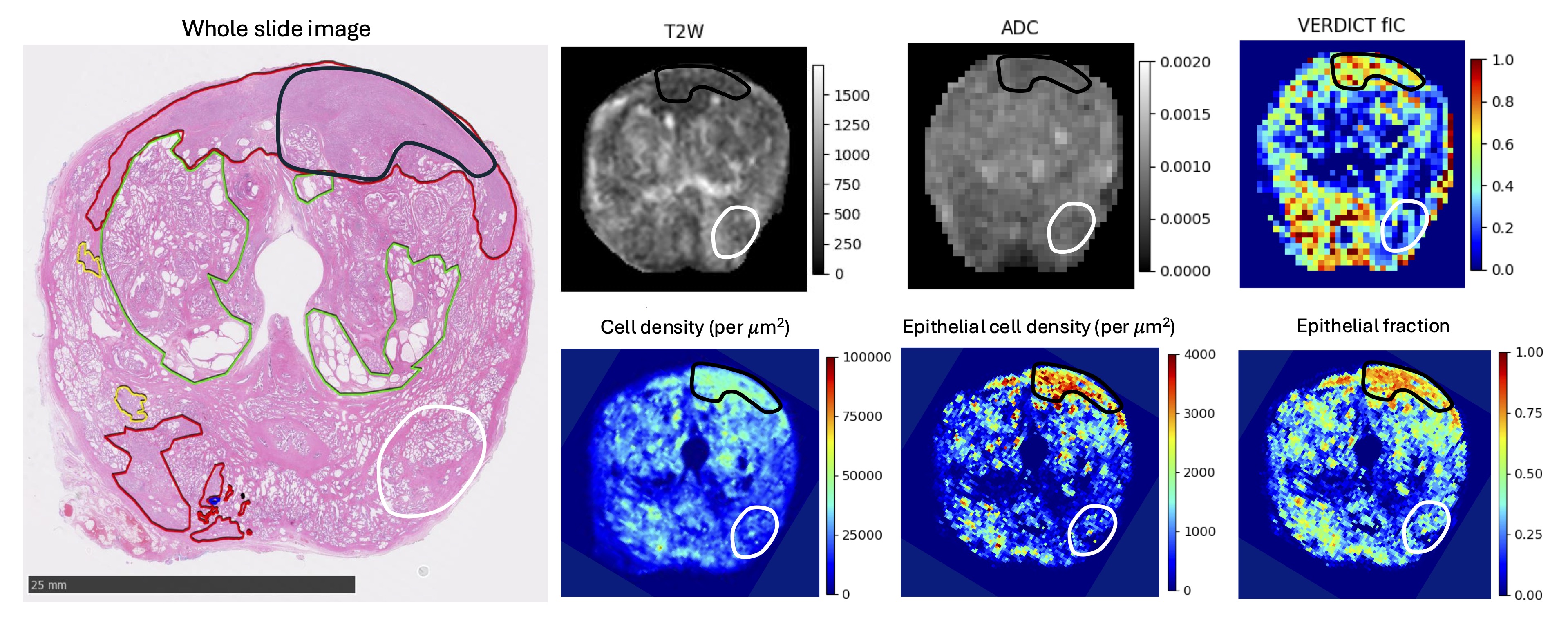
More recent papers have further demonstrated the usability of VERDICT-MRI derived maps. Singh et al. showed in 2022 that fIC maps were better suited for classification into csPCa and non-csPCa than ADC and PSA density levels [3]. In particular, it was the only one that was able to discriminate lesions that scored 3 on the Likert scale. Sen et al. then showed that model-based methods such as VERDICT-MRI produced significant differences between false positives and normal tissue [9]. The in-vivo maps were also validated by comparison to volume fractions derived from histology images [10], after visual co-registration. Building on this histological validation work, Masramon et al. (2025) used personalised 3D-printed molds and whole-mount prostatectomy specimens to rigorously validate the biological origin of fIC, demonstrating strong correlations between in-vivo VERDICT fIC, epithelial fraction and histological cell density, and thereby reinforcing fIC as a biologically interpretable marker of tumour cellularity [11]. Singh et al. (2023) compared image quality of conventional ADC maps and VERDICT-derived fIC maps using the PI-QUAL score and a dedicated artefact Likert scale, showing that fIC maps achieve at least comparable image quality while offering superior lesion conspicuity and diagnostic performance [12]. Finally, parameter maps from a new version of the VERDICT model (rVERDICT) outperformed the standard VERDICT-MRI and the ADC maps in the task of Gleason grade discrimination (between 3+3, 3+4 and ≥ 3 + 4) [13].
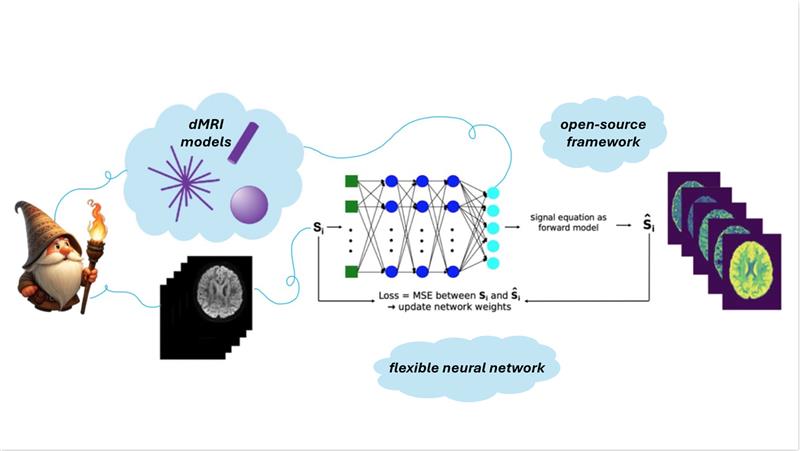
Model fitting
Sen et al. introduced ssVERDICT, a self-supervised deep learning approach for fitting the VERDICT model that removes the need for explicit training labels and outperforms both conventional non-linear least squares and supervised deep learning fits in simulations, while improving lesion conspicuity and benign–malignant discrimination in vivo [14]. Sen et al. also created an open-source PyTorch toolkit (microTorch) implementing VERDICT and other microstructural MRI models, supporting simulation, fitting, and deep-learning-based model estimation [15].
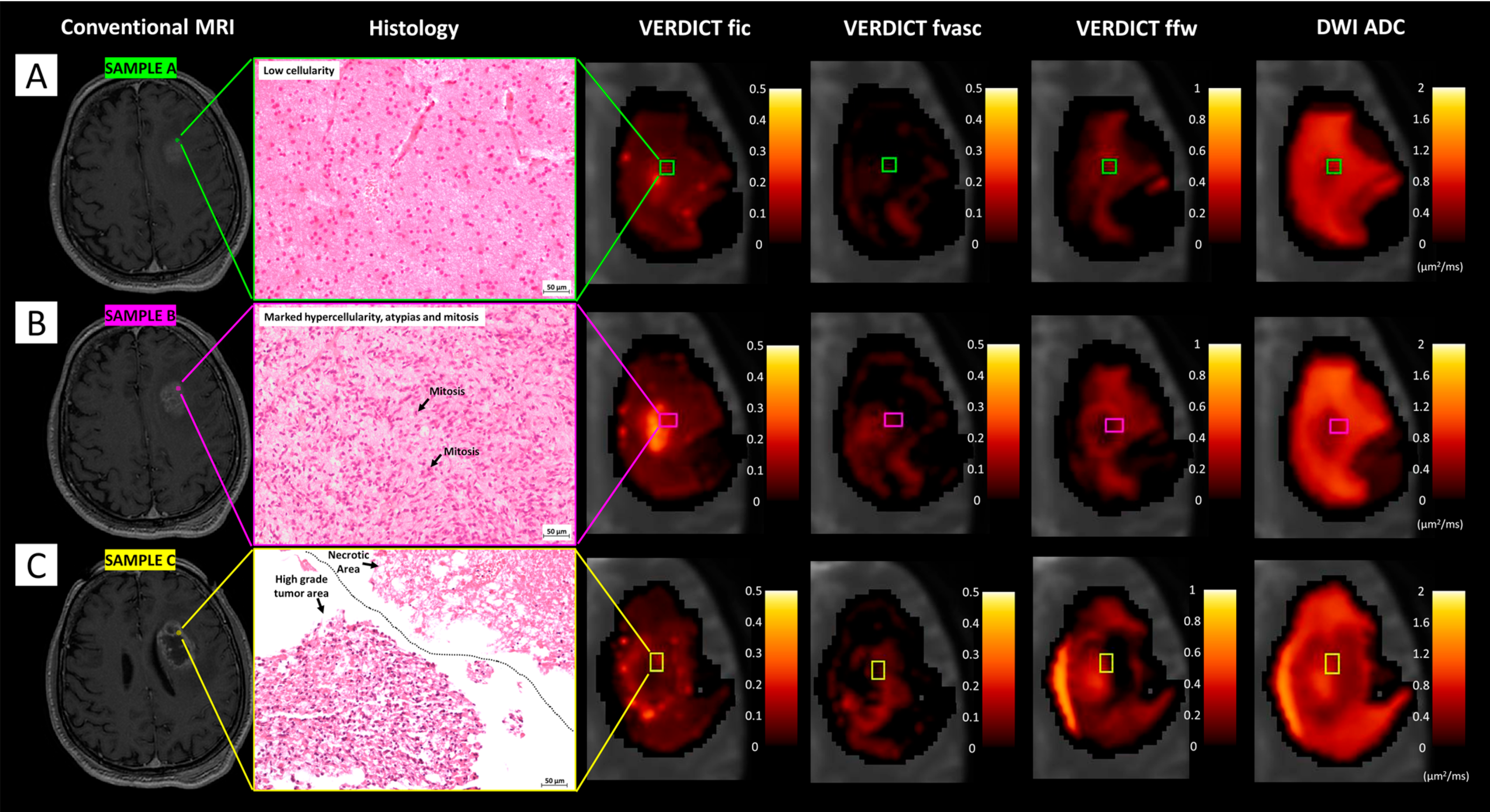
VERDICT-MRI in other organs
Beyond the prostate, the VERDICT framework has also been extended to other tumour sites. Figini et al. applied VERDICT-MRI to brain tumours, demonstrating that VERDICT-derived cellular and vascular measures are feasible in the brain and show good agreement with histology, thereby supporting VERDICT as a general diffusion-based framework for tumour microstructural characterisation [16]. Sen et al. further extended VERDICT-MRI to renal tumours using a dual deep learning approach: a self-supervised network for VERDICT model fitting and a second network for non-invasive renal tumour subtyping. They showed that a three-compartment renal VERDICT model fits the diffusion data more accurately than IVIM or ADC, and that VERDICT-derived parameters can discriminate between different renal tumour subtypes while also enabling optimisation of shorter acquisition protocols[17].
Further Reading
[1] Eleftheria Panagiotaki, Rachel W Chan, Nikolaos Dikaios, Hashim U Ahmed, James O’callaghan, Alex Freeman, David Atkinson, Shonit Punwani, David J Hawkes, and Daniel C Alexander. Microstructural characterization of normal and malignant human prostate tissue with vascular, extracellular, and restricted diffusion for cytometry in tumours magnetic resonance imaging. 2015.
[2] Edward W. Johnston, Elisenda Bonet-Carne, Uran Ferizi, Ben Yvernault, Hayley Pye, Dominic Patel, Joey Clemente, Wivijin Piga, Susan Heavey, Harbir S. Sidhu, Francesco Giganti, James O’Callaghan, Mrishta Brizmohun Appayya, Alistair Grey, Alexandra Saborowska, Sebastien Ourselin, David Hawkes, Car- oline M. Moore, Mark Emberton, Hashim U. Ahmed, Hayley Whitaker, Manuel Rodriguez-Justo, Alexander Freeman, David Atkinson, Daniel Alexander, Eleft- heria Panagiotaki, and Shonit Punwani. Verdict mri for prostate cancer: Intracellular volume fraction versus apparent diffusion coefficient. Radiology, 291:391–397, 5 2019.
[3] Saurabh Singh, Harriet Rogers, Baris Kanber, Joey Clemente, Hayley Pye, Edward W. Johnston, Tom Parry, Alistair Grey, Eoin Dinneen, Greg Shaw, Susan Heavey, Urszula Stopka-Farooqui, Aiman Haider, Alex Freeman, Francesco Giganti, David Atkinson, Caroline M. Moore, Hayley C. Whitaker, Daniel C. Alexander, Eleftheria Panagiotaki, and Shonit Punwani. Avoiding unnecessary biopsy after multiparametric prostate mri with verdict analysis: The innovate study. Radiology, 8 2022.
[4] Saurabh Singh, Manju Mathew, Thomy Mertzanidou, Shipra Suman, Joey Clemente, Adam Retter, Marianthi Vasiliki Papoutsaki, and Lorna Smith et al. Histo-mri map study protocol: a prospective cohort study mapping mri to histology for biomarker validation and prediction of prostate cancer. BMJ open, 12:e059847, 4 2022.
[5] Elisenda Bonet-Carne, Maira Tariq, Hayley Pye, Brizmohun Appayya, Aiman Haider, Colleen Bayley, Joseph Jacobs, Alexander Freeman, David Hawkes, David Atkinson, Greg Shaw, Hayley Whitaker, Daniel C Alexander, Shonit Punwani, and Eleftheria Panagiotaki. Histological validation of in-vivo verdict mri for prostate using 3d personalised moulds. 2017.
[6] Mrishta Brizmohun Appayya, Edward W Johnston, Arash Latifoltojar, James O’callaghan, Elisenda Bonnet-Carne, Hayley Pye, Dominic Patel, Susan Heavey, Alistair Grey, Sebastien Ourselin, David Hawkes, Caroline Moore, Hay- ley Whitaker, Alexander Freeman, David Atkinson, Daniel Alexander, Elefthe- ria Panagiotaki, and Shonit Punwani. The intracellular component of verdict (vascular, extracellular, and restricted diffusion for cytometry in tumors) mri distinguishes gleason 4 pattern better than apparent diffusion coefficient. 2018.
[7] Eleni Chiou, Francesco Giganti, Elisenda Bonet-Carne, Shonit Punwani, Iasonas Kokkinos, and Eleftheria Panagiotaki. Prostate cancer classification on verdict dw-mri using convolutional neural networks. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), 11046 LNCS:319–327, 2018.
[8] Colleen Bailey, Roger M. Bourne, Bernard Siow, Edward W. Johnston, Mrishta Brizmohun Appayya, Hayley Pye, Susan Heavey, Thomy Mertzanidou, Hayley Whitaker, Alex Freeman, Dominic Patel, Greg L. Shaw, Ashwin Sridhar, David J. Hawkes, Shonit Punwani, Daniel C. Alexander, and Eleftheria Panagiotaki. Verdict mri validation in fresh and fixed prostate specimens using patient-specific moulds for histological and mr alignment. NMR in Biomedicine, 32, 5 2019.
[9] Snigdha Sen, Vanya Valindria, Paddy J. Slator, Hayley Pye, Alistair Grey, Alex Freeman, Caroline Moore, Hayley Whitaker, Shonit Punwani, Saurabh Singh, and Eleftheria Panagiotaki. Differentiating false positive lesions from clinically significant cancer and normal prostate tissue using verdict mri and other diffusion models. Diagnostics, 12, 7 2022.
[10] Aritrick Chatterjee, Crystal Mercado, Roger M. Bourne, Ambereen Yousuf, Brittany Hess, Tatjana Antic, Scott Eggener, Aytekin Oto, and Gregory S. Karczmar. Validation of prostate tissue composition by using hybrid multidi- mensional mri: Correlation with histologic findings. Radiology, 302:368–377, 2 2022.
[11] Marta Masramon, Manju Mathew, Saurabh Singh, Thomy Mertzanidou, Joey Clemente, Adam Retter, Natasha Thorley, Adam Phipps, Thomas Parry, Marianthi-Vasiliki Papoutsaki, Lorna Smith, Francesco Grussu, Veeru Kasivisvanathan, Alistair Grey, Eoin Dinneen, Greg Shaw, Dominic Patel, Lucy Caselton, Caroline M. Moore, David Atkinson, Aiman Haider, Alex Freeman, Daniel C. Alexander, Shonit Punwani, and Eleftheria Panagiotaki. Validating the biological origin of in vivo fractional intracellular volume from VERDICT-MRI in the prostate. Research Square preprint, 2025.
[12] Snigdha Sen, Lorna Smith, Lucy Caselton, Joey Clemente, Minh Tran, Shonit Punwani, Eleftheria Panagiotaki. Dual Deep Learning Approach for Non-invasive Renal Tumour Subtyping with VERDICT-MRI. arXiv:2504.07246, 2025.
[13] Marco Palombo, Vanya Valindria, Saurabh Singh, Eleni Chiou, Francesco Gi- ganti, Hayley Pye, Hayley C. Whitaker, David Atkinson, Shonit Punwani, Daniel C. Alexander, and Eleftheria Panagiotaki. Joint estimation of relaxation and diffusion tissue parameters for prostate cancer with relaxation-verdict mri. Scientific Reports, 13, 12 2023.
[14] Snigdha Sen. microTorchFit: PyTorch implementations of microstructural and quantitative MRI models. GitHub repository, 2025. Available at: https://github.com/snigdha-sen/microtorchfit
[15] Saurabh Singh, Francesco Giganti, Louise Dickinson, Harriet Rogers, Baris Kanber, Joey Clemente, Hayley Pye, Susan Heavey, Urszula Stopka-Farooqui, Edward W. Johnston, Caroline M. Moore, Alex Freeman, Hayley C. Whitaker, Daniel C. Alexander, Eleftheria Panagiotaki, and Shonit Punwani. Prostate MR image quality of ADC maps versus fIC maps from VERDICT MRI using PI-QUAL and a dedicated Likert artefact score. European Journal of Radiology, 2023.
[16] Snigdha Sen, Saurabh Singh, Hayley Pye, Caroline M. Moore, Hayley Whitaker, Shonit Punwani, David Atkinson, Eleftheria Panagiotaki, and Paddy J. Slator. ssVERDICT: Self-Supervised VERDICT-MRI for Enhanced Prostate Tumour Characterisation. arXiv:2309.06268, 2023.
[17] Matteo Figini, Antonella Castellano, Michele Bailo, Marcella Callea, Marcello Cadioli, Samira Bouyagoub, Marco Palombo, Valentina Pieri, Pietro Mortini, Andrea Falini, Daniel C. Alexander, Mara Cercignani, and Eleftheria Panagiotaki. Comprehensive Brain Tumour Characterisation with VERDICT-MRI: Evaluation of Cellular and Vascular Measures Validated by Histology. Cancers, 2023.
